Kaggle: Predict the occurrence of diabetes
This project presents a code/kernel used in a Kaggle competition promoted by Data Science Academy.
This project presents a code/kernel used in a Kaggle competition promoted by Data Science Academy in January of 2019.
The goal of the competition was to create a Machine Learning model to predict the occurrence of diabetes.
Data source: National Institute of Diabetes and Digestive and Kidney Diseases
Competition page: kaggle.com/c/competicao-dsa-machine-learnin..
Predict the occurrence of diabetes
Above the EDA is presented with the source code used to perform the data pre-processing, data transformation, and create the machine learning models.
Exploratory Data Analysis
Data fields:
- num_gestacoes - Number of times pregnant
- glicose - Plasma glucose concentration in oral glucose tolerance test
- pressao_sanguinea - Diastolic blood pressure in mm Hg
- grossura_pele - Thickness of the triceps skinfold in mm
- insulina - Insulin (mu U / ml)
- bmi - Body mass index measured by weight in kg / (height in m) ^ 2
- indice_historico - Diabetes History Index (Pedigree Function)
- idade - Age in years
- classe - Class (0 - did not develop disease / 1 - developed disease)
Loading the data
# Importing packages
import numpy as np
import pandas as pd
import seaborn as sns
from scipy import stats
import matplotlib.pyplot as plt
import warnings
warnings.filterwarnings("ignore", category=FutureWarning)
%matplotlib inline
# Loading the data
data = pd.read_csv('data/dataset_treino.csv')
test_data = pd.read_csv('data/dataset_teste.csv')
data.head(5)
| id | num_gestacoes | glicose | pressao_sanguinea | grossura_pele | insulina | bmi | indice_historico | idade | classe | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 6 | 148 | 72 | 35 | 0 | 33.6 | 0.627 | 50 | 1 |
| 1 | 2 | 1 | 85 | 66 | 29 | 0 | 26.6 | 0.351 | 31 | 0 |
| 2 | 3 | 8 | 183 | 64 | 0 | 0 | 23.3 | 0.672 | 32 | 1 |
| 3 | 4 | 1 | 89 | 66 | 23 | 94 | 28.1 | 0.167 | 21 | 0 |
| 4 | 5 | 0 | 137 | 40 | 35 | 168 | 43.1 | 2.288 | 33 | 1 |
Data overview
# General statistics
data.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 600 entries, 0 to 599
Data columns (total 10 columns):
id 600 non-null int64
num_gestacoes 600 non-null int64
glicose 600 non-null int64
pressao_sanguinea 600 non-null int64
grossura_pele 600 non-null int64
insulina 600 non-null int64
bmi 600 non-null float64
indice_historico 600 non-null float64
idade 600 non-null int64
classe 600 non-null int64
dtypes: float64(2), int64(8)
memory usage: 47.0 KB
All 10 predictors variables (features) are quantitative (numerical) and we have 600 observations to build the prediction model.
The only qualitative column is the labels, where:
- 0 - do not have the disease
- 1 - have the disease
Data Cleaning
Checking if there are missing values
# If the result is False, there is no missing value
data.isnull().values.any()
False
Computing statistics for each column
data.describe()
| id | num_gestacoes | glicose | pressao_sanguinea | grossura_pele | insulina | bmi | indice_historico | idade | classe | |
|---|---|---|---|---|---|---|---|---|---|---|
| count | 600.000000 | 600.000000 | 600.000000 | 600.000000 | 600.000000 | 600.000000 | 600.000000 | 600.000000 | 600.000000 | 600.000000 |
| mean | 300.500000 | 3.820000 | 120.135000 | 68.681667 | 20.558333 | 79.528333 | 31.905333 | 0.481063 | 33.278333 | 0.346667 |
| std | 173.349358 | 3.362009 | 32.658246 | 19.360226 | 16.004588 | 116.490583 | 8.009638 | 0.337284 | 11.822315 | 0.476306 |
| min | 1.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.078000 | 21.000000 | 0.000000 |
| 25% | 150.750000 | 1.000000 | 99.000000 | 64.000000 | 0.000000 | 0.000000 | 27.075000 | 0.248000 | 24.000000 | 0.000000 |
| 50% | 300.500000 | 3.000000 | 116.000000 | 70.000000 | 23.000000 | 36.500000 | 32.000000 | 0.384000 | 29.000000 | 0.000000 |
| 75% | 450.250000 | 6.000000 | 140.000000 | 80.000000 | 32.000000 | 122.750000 | 36.525000 | 0.647000 | 40.000000 | 1.000000 |
| max | 600.000000 | 17.000000 | 198.000000 | 122.000000 | 99.000000 | 846.000000 | 67.100000 | 2.420000 | 81.000000 | 1.000000 |
From the table above, we can see the zero values in almost all columns. For some of these columns, zero makes sense, like for Pregnancies and Outcome. But for some of the others, like BloodPressure or BMI, zero definitely doesn't make sense.
After read some papers about the variables in the dataset, I see that some columns can have a value very close to zero (e.g. grossura_pele), but others can't have a zero value.
The columns can not have a zero value.
- glicose
- pressao_sanguinea
- bmi
Let's see the number of the occurrences of zero values for all columns:
# Compute the number of occurrences of a zero value
features = ['num_gestacoes', 'glicose', 'pressao_sanguinea', 'grossura_pele', 'insulina', 'bmi', 'indice_historico', 'idade']
for c in features:
counter = len(data[data[c] == 0])
print('{} - {}'.format(c, counter))
num_gestacoes - 93
glicose - 5
pressao_sanguinea - 28
grossura_pele - 175
insulina - 289
bmi - 9
indice_historico - 0
idade - 0
We can also see that column insulina has 289 values, which correspond to 48% of the training data.
Let's remove these observations from the selected columns.
# Removing observations with zero value
data_cleaned = data.copy()
for c in ['glicose', 'pressao_sanguinea', 'bmi']:
data_cleaned = data_cleaned[data_cleaned[c] != 0]
data_cleaned.shape
(564, 10)
The final number of observations was 564.
Let's see the compute some statistics again:
data_cleaned.describe()
| id | num_gestacoes | glicose | pressao_sanguinea | grossura_pele | insulina | bmi | indice_historico | idade | classe | |
|---|---|---|---|---|---|---|---|---|---|---|
| count | 564.000000 | 564.000000 | 564.000000 | 564.000000 | 564.000000 | 564.000000 | 564.000000 | 564.000000 | 564.000000 | 564.000000 |
| mean | 300.664894 | 3.845745 | 121.354610 | 72.049645 | 21.432624 | 84.406028 | 32.367199 | 0.483294 | 33.448582 | 0.340426 |
| std | 173.410435 | 3.349287 | 31.130992 | 12.261552 | 15.809953 | 118.432015 | 6.974710 | 0.337668 | 11.868844 | 0.474273 |
| min | 1.000000 | 0.000000 | 44.000000 | 24.000000 | 0.000000 | 0.000000 | 18.200000 | 0.078000 | 21.000000 | 0.000000 |
| 25% | 150.750000 | 1.000000 | 99.000000 | 64.000000 | 0.000000 | 0.000000 | 27.300000 | 0.250500 | 24.000000 | 0.000000 |
| 50% | 298.500000 | 3.000000 | 116.000000 | 72.000000 | 23.500000 | 49.000000 | 32.000000 | 0.389000 | 29.000000 | 0.000000 |
| 75% | 450.250000 | 6.000000 | 141.250000 | 80.000000 | 33.000000 | 130.000000 | 36.600000 | 0.648250 | 41.000000 | 1.000000 |
| max | 600.000000 | 17.000000 | 198.000000 | 122.000000 | 99.000000 | 846.000000 | 67.100000 | 2.420000 | 81.000000 | 1.000000 |
Checking outliers
Let's check if there are outliers in the data.
First, we'll use a set of boxplots, one for each column.
fig, axes = plt.subplots(2,4, figsize=(20,8))
x,y = 0,0
for i, column in enumerate(data_cleaned.columns[1:-1]):
sns.boxplot(x=data_cleaned[column], ax=axes[x,y])
if i < 3:
y += 1
elif i == 3:
x = 1
y = 0
else:
y += 1
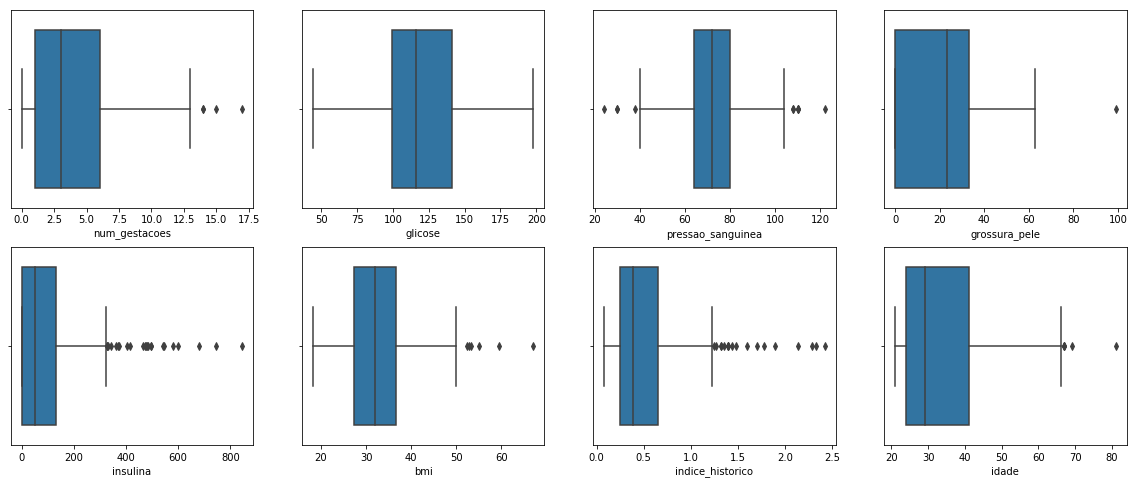
We can see some possible outliers for almost all columns (separated points in the plots).
The outliers can either be a mistake or just variance. For now, let's consider all of them as mistakes.
To remove these outliers we can use Z-Score or IQR (Interquartile Range).
The Z-score is the signed number of standard deviations by which the value of an observation or data point is above the mean value of what is being observed or measured. Z-score is finding the distribution of data where the mean is 0 and the standard deviation is 1. While calculating the Z-score we re-scale and center the data and look for data points that are too far from zero. These data points which are way too far from zero will be treated as outliers. In most of the cases, a threshold of 3 or -3 is used i.e if the Z-score value is greater than or less than 3 or -3 respectively, that data point will be identified as outliers.
Let's use the Z-score function defined in the Scipy library to detect the outliers:
# Compute the Z-Score for each column
print(data_cleaned.shape)
z = np.abs(stats.zscore(data_cleaned))
data_cleaned = data_cleaned[(z < 3).all(axis=1)]
print(data_cleaned.shape)
(564, 10)
(531, 10)
Using Z-Score, 33 observations were removed.
Let's see the boxplots again:
fig, axes = plt.subplots(2,4, figsize=(20,8))
x,y = 0,0
for i, column in enumerate(data_cleaned.columns[1:-1]):
sns.boxplot(x=data_cleaned[column], ax=axes[x,y], palette="Set2")
if i < 3:
y += 1
elif i == 3:
x = 1
y = 0
else:
y += 1
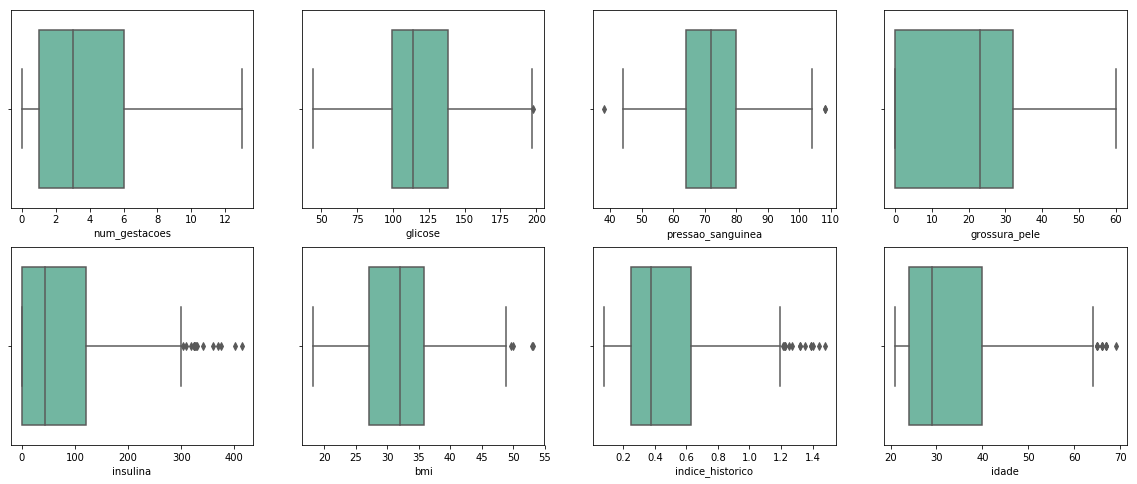
The data was much cleaner now. Still, there are some points in the boxplots, but some of them are not outliers, like the insulina values higher than 400, which is acceptable in people with diabetes.
Checking the balance of the dataset
Let's checking the distribuitions of examples for each label:
data_cleaned.classe.value_counts().plot(kind='bar');
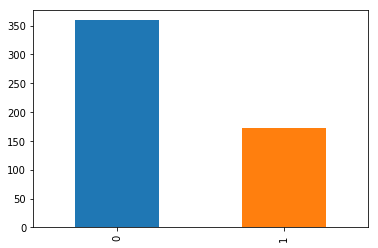
data_cleaned.classe.value_counts(normalize=True)
0 0.676083
1 0.323917
Name: classe, dtype: float64
From the figure above, we see most of our examples are of people that do not have the disease. More specifically, 67% of the data are for healthy people.
As the dataset is unbalanced, let's use some methods to reduce the unbalance of the classes.
I use the over-sampling SMOTE method. SMOTE (Synthetic Minority Oversampling Technique) consists of synthesizing elements for the minority class, based on those that already exist. It works randomly picking a point from the minority class and computing the k-nearest neighbors for this point. The synthetic points are added between the chosen point and its neighbors.
from imblearn.over_sampling import SMOTE
# Select the columns with features
features = ['num_gestacoes', 'glicose', 'pressao_sanguinea', 'grossura_pele', 'insulina', 'bmi', 'indice_historico', 'idade']
X = data_cleaned[features]
# Select the columns with labels
Y = data_cleaned['classe']
smote = SMOTE(sampling_strategy=1.0, k_neighbors=4)
X_sm, y_sm = smote.fit_sample(X, Y)
print(X_sm.shape[0] - X.shape[0], 'new random picked points')
data_cleaned_oversampled = pd.DataFrame(X_sm, columns=data.columns[1:-1])
data_cleaned_oversampled['classe'] = y_sm
data_cleaned_oversampled['id'] = range(1,len(y_sm)+1)
for c in ['num_gestacoes', 'glicose', 'pressao_sanguinea', 'grossura_pele', 'insulina', 'idade']:
data_cleaned_oversampled[c] = data_cleaned_oversampled[c].apply(lambda x: int(x))
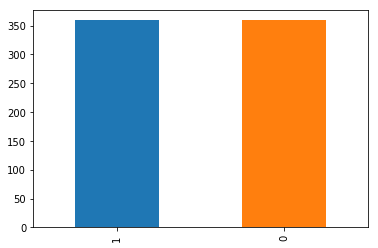
data_cleaned_oversampled.classe.value_counts().plot(kind='bar');
187 new random picked points
Training the Model
Now that data is cleaned, let's use a machine learning model to predict whether or not a person has diabetes.
The metric used in this competition is the accuracy score.
Using Decision Tree
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score
from sklearn.metrics import confusion_matrix
from sklearn import tree
# Select the columns with features
features = ['num_gestacoes', 'glicose', 'pressao_sanguinea', 'grossura_pele', 'insulina', 'bmi', 'indice_historico', 'idade']
X = data_cleaned_oversampled[features]
# Select the columns with labels
Y = data_cleaned_oversampled['classe']
# Perform the training and test 100 times with different seeds and compute the mean accuracy.
# Save results
acurrances = []
for i in range(100):
# Spliting Dataset into Test and Train
X_train, X_test, y_train, y_test = train_test_split(X, Y, test_size=0.3, random_state=i)
# Create and train the model
clf = tree.DecisionTreeClassifier(criterion = 'entropy', random_state=i, max_depth=4)
clf.fit(X_train,y_train)
# Performing predictions with test dataset
y_pred = clf.predict(X_test)
# Computing accuracy
acurrances.append(accuracy_score(y_test, y_pred)*100)
print('Accuracy is ', np.mean(acurrances))
Accuracy is 73.35648148148148
Using Logistic Regression (LR)
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import classification_report, confusion_matrix
# Select the columns with features
features = ['num_gestacoes', 'glicose', 'pressao_sanguinea', 'grossura_pele', 'insulina', 'bmi', 'indice_historico', 'idade']
# For the LR the oversampled database decreased the model accuracy, so I choose do not to use it.
X = data_cleaned[features]
# Select the columns with labels
Y = data_cleaned['classe']
# Perform the training and test 100 times with different seeds and compute the mean accuracy.
# Save results
acurrances = []
for i in range(100):
# Spliting the data
X_train, X_test, y_train, y_test = train_test_split(X, Y, test_size=0.3, random_state=i)
LR_model=LogisticRegression(class_weight={1:1.15})
LR_model.fit(X_train,y_train)
# Testing
y_pred=LR_model.predict(X_test)
acurrances.append(accuracy_score(y_test, y_pred)*100)
# Print only the last
if i == 99:
pass
#print(classification_report(y_test,y_pred))
#print(confusion_matrix(y_true=y_test, y_pred=y_pred))
print('Accuracy is ', np.mean(acurrances))
Accuracy is 76.725
Feature selection
The large number of zero values in the grossura_pele and insulina are impairing the performance of the model. So, as insulin is an important parameter in the evaluation of diabetes let's replace the zeros values by the mean for both columns.
As the Logistic Regression (LR) performs better than the Decision Tree. I'll use only LR from now.
# Replacing the zeros by the mean
data_cleaned_no_zeros = data_cleaned.copy()
for c in ['grossura_pele', 'insulina']:
feature_avg =data_cleaned[data_cleaned[c]>0][[c]].mean()
data_cleaned[c]=np.where(data_cleaned[[c]]!=0,data_cleaned[[c]],feature_avg)
# Select the columns with features
features = ['num_gestacoes', 'glicose', 'pressao_sanguinea', 'grossura_pele', 'insulina', 'bmi', 'indice_historico', 'idade']
X = data_cleaned_no_zeros[features]
# Select the columns with labels
Y = data_cleaned_no_zeros['classe']
# Perform the training and test 100 times with different seeds and compute the mean accuracy score.
# Save results
acurrances = []
for i in range(100):
# Spliting the data
X_train, X_test, y_train, y_test = train_test_split(X, Y, test_size=0.3, random_state=i)
LR_model=LogisticRegression(class_weight={1:1.1})
LR_model.fit(X_train,y_train)
# Testing
y_pred=LR_model.predict(X_test)
acurrances.append(accuracy_score(y_test, y_pred)*100)
# Print only the last
if i == 99:
print(classification_report(y_test,y_pred))
print(confusion_matrix(y_true=y_test, y_pred=y_pred))
print('Accuracy is ', np.mean(acurrances))
precision recall f1-score support
0 0.83 0.91 0.87 110
1 0.75 0.60 0.67 50
micro avg 0.81 0.81 0.81 160
macro avg 0.79 0.75 0.77 160
weighted avg 0.81 0.81 0.81 160
[[100 10]
[ 20 30]]
Accuracy is 76.5625
The performance decrease when I replace the zeros values by the mean for grossura_pele and insulina columns.
Performing the Final Prediction
For the final model, I choose to use LR with the cleaned dataset.
# Create and train the model with all data
model=LogisticRegression(class_weight={1:1.1})
model.fit(data_cleaned[features],data_cleaned['classe'])
# Get the kaggle test data
X_test = test_data[features]
# Make the prediction
prediction = model.predict(X_test)
# Add the predictions to the dataframe
test_data['classe'] = prediction
# Create the submission file
test_data.loc[:,['id', 'classe']].to_csv('submission.csv', encoding='utf-8', index=False)
Source code
The solution is also available at Github.
How to use
- You will need Python 3.5+ to run the code.
- Python can be downloaded here.
- You have to install some Python packages, in command prompt/Terminal:
pip install jupyter-lab scikit-learn pandas seaborn matplotlib Once you have installed the required packages, just clone/download this project:
git clone https://github.com/cpatrickalves/kaggle-diabetes-predictionAccess the project folder in command prompt/Terminal and run the following command:
jupyter-labThen open the kernel file.

